shu251@uw.edu
//Vt// data google doc
August 16, 2010
Still analyzing data from qPCR runs with Vibrio tubiashii in different pCO2 conditions and anemones challenged with Vt.
Vt virulence expression seems to increase from 4 to 24 hours post inoculation.
Vt virulence gene expression also seems to be higher in anemones with heat shock and Vt challenge. This may be due to heat shock providing a better medium for the Vt to colonize or it may be due to the anemone being more vulnerable, this allowing Vt to express more virulence.
More to come!
No expression from Littorina samples
August 15, 2010
Analyzing data from qPCR
Have data from V. tubiashii in two pCO2 conditions.
V. tubiashii from anemones heat stressed and not heat stressed.
Without normalizing gene, I can only rely on the fact that I extracted approximately the same volume from all my samples (from pCO2 conditions). Based on consistent volumes used for all subsequent reactions and procedures, I can use a standard/ consistent threshold to compare my results.
Average efficiency
Ro=[1/(1+AE)^ct]
August 14, 2010
Timeline for today:
9AM: inoculate
Before lunch, maybe extract RNA, make cDNA for two samples? from elevated CO2 from previous experiment
After lunch (12:30-1:00PM): take 4 hour sample
Extraction RNA from filtered water (approx 100-300mLs), will have 8 samples
Make cDNA (8 samples total)
Run qPCR at night (8 samples with 2 genes: chitinase and vtpR)
9PM: take 12 hour sample (6 samples)
*In the interest of time and to not change densities of Emma's experiment.... may be taking 100 mL at 12 hours for filtration and then get 200+mLs at 24 hours to filter
Design:
Before lunch today: RNA extraction for elevated CO2 and possibly 2 hours post inoculation?
pCO2s for previous experiment (2-3 time points)
take samples from Emma's: 4-6 hours post inoculation, 12 hours (with oyster) and 24 hours (a whole litre)
RNA extractions: C840 from 8/5 and 8/7
Will be running qPCR plate tonight with all genes (chitinase, VtpR, VtpA)
plate will have controls from 4 hour and 24 hours at ambient and elevated pCO2.
Will also include samples from Loni and Marie's anemone's exposed to Vt (control with no Vt and then animal with Vt).
qPCR will examine V. tubiashii in current pCO2 conditions and elevated conditions.
Virulence genes that are potentially influenced by pH changes in the environment have larger implications for host-pathogen relationship amidst climatic changes. The next step from here would be to examine virulence gene expression with and without a host present. Have been trying to use oyster larvae, but will now look at a couple of samples from anemone exposed to Vt.
New plans:
Although last minute, the scramble for data has begun. No oyster larvae were found today, thus the second experiment is delayed to probably after the class. I will be going back original experimental design.
Objectives:
1. Vibrio tubiashii in two CO2 environments (380ppm and 840 ppm)
2. Vibrio tubiashii in several species: anemone, snail, and oyster larvae.
From these two perspectives I will be examining potential differences in virulent associated genes.
Three phases:
qPCR Saturday (2pm): with controls of Vt from different pCO2s, also included are the anemone
Anemone were inoculated with 2 mLs of Vt (concentration pending...)
They sat for 3 hours and then tentacles were taken.
cDNA will be run for control (no Vt), treatment1- Vt, and treatment2- Vt+heat shock
August 13, 2010
Running a conventional PCR today to screen my primers to see if they even work. Last night, ran the qPCR with 16S normalizing and got no gene expression. Annealing temperature was lowered to 50C and cDNA was transcribed from random primers.
PCR today consists of 8/6 samples (24hours), from container A at 380ppm and the control that corresponds. I also included the cultured Vt as a possible positive control. Will hopefully get bands later and then qPCRs will be more productive.
Conventional PCR:
Reagents/reaction:
- 12.5uL of mix from differential display kit (includes dye, buffer, etc)
- 0.8uL primer F
- 0.8uL primer R
- 8.9uL of PCR water
Used 4 primers and did samples: control Vt, control 380, A380, A380oyster larvae, and blank
Primers: 16S, chitinase, VtpA, VtpR
Gel:
Ran at 110V for 20 minutes and got bands with Chitinase and VtpR primers, 16S and VtpA did not see any banding patterns.
Still concerned with Vt collection and getting results!
Started qPCR:
Running chitinase and VtpR
Planning experiment start for tomorrow:
- Will be inoculating water/larvae at approx. 9AM.
- Take 4 hour, 12 hour, and 24 hours samples.
- after 4th hour samples are taken, begin RNA extraction and make cDNA, hopefully can do qPCR right away.
- can probably do qPCR right around dinner time tomorrow with controls from 840ppm from previous experiment and from 4 hour samples... then compare 12 and 24 hour samples on Sunday, hopefully will have sufficient data to present this next week
Timeline for tomorrow:
9AM: inoculate
Before lunch, maybe extract RNA, make cDNA for two samples? from elevated CO2 from previous experiment
After lunch (12:30-1:00PM): take 4 hour sample
Extraction RNA from filtered water (approx 100-300mLs), will have 8 samples
Make cDNA (8 samples total)
Run qPCR at night (8 samples with 2 genes: chitinase and vtpR)
9PM: take 12 hour sample (6 samples)
Sunday: take 24 hour sample
Extraction RNA (12 samples), cDNA, qPCR
Data!
August 12, 2010
Making cDNA:
- Jackson helped aliquot reagents for reverse transcription to make cDNA
- RTing samples from 8/5 and 8/6 under ambient CO2 conditions (380ppm)
- Planning on running qPCR tonight with 16S normalizing
| 5 (8/5) |
6 (8/6) |
|
| A |
C |
C |
| B |
C |
C |
| C |
A |
A |
| D |
A |
A |
| E |
L |
L |
| F |
L |
L |
| G |
Vt |
blank |
| F |
Vt |
blank |
Thermal profile:
- 7 minutes, 95C
- 10 seconds, 95C
- 30 seconds, 50C (trial at 50C, maybe use 55C if it doesn't work)
- 30 seconds, 72C
- 1 minutes, 95C
- 30 seconds, 55C
- 30 seconds, 95C
- repeat for 40 cycles (from step 2)
- qPCR started at 7:45pm
- put Vt on plate for Elene
- if qPCR goes well, do more cDNA tonight?
- next steps for qPCR?
August 11, 2010
Issue:
Because I'm working with prokaryotes, they don't have a poly A tail! So the oligo dT primers will not work.
Which is unfortunate. When making cDNA from RNA, you can use two other types of primers, use a sequence specific primer for the RNA or use random primers. Random primers are hopefully on their way.
- Focusing today on getting data from Loni and Marie's anemone inoculated samples and try to compare with oyster exposed.
- make marine agar plates
- grow up Vt
- Another plan of attack with my RNA from Vt?
Timeline:
- primers received by Friday, cDNA 12-15 samples
- run qPCR for normalizing gene
- run qPCR for other three genes (time permitting, separately)
- analyze!
Oyster collection
- Collected for spawning
- helping Emma start a new experiment
- Experiment will begin and hopefully inoculation will begin Friday or this weekend
Next steps:
Hopefully I will get the random primers I can use in the RNA to cDNA for Vt.
As soon as those come in I can complete the reverse transcription to make cDNA.
Try 16S gene for normalizing and then go from there.
August 10, 2010
- Continue work on proposal/Quiz#3
- Extracted more RNA last night
- Before lunch: make cDNA, and prep qPCR
- Collect oyster samples and count mortality, etc.
cDNA samples:
In PCR plate:
Added 6uL of RNA and brought total volume up to 17.75 uL
added 0.5uL of Oligo dT primers
Heat at 70C for 5 minutes and then ice for 10 minutes
Add master mix for cDNA:
Ingredients:
- 5uL 5x buffer
- 1.25uL 10mM dNTPs
- 0.5uL reverse transcriptase
- had 8 reactions to do today, made cDNA master mix for 9 reactions
Cycle: 42C for 1 hour and 95C for 3 minutes
- store at -20C
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
| A |
C380. 8/5 |
C380. 8/6 |
C380. 8/7 |
A oys 8/7 |
||||
| B |
C380. 8/5 |
C380. 8/6 |
C380. 8/7 |
A oys 8/7 |
||||
| C |
A 8/5 |
A 8/6 |
A 8/7 |
L oys 8/7 |
||||
| D |
A 8/5 |
A 8/6 |
A 8/7 |
L oys 8/7 |
||||
| E |
L 8/5 |
L 8/6 |
L 8/7 |
blank |
||||
| F |
L 8/5 |
L 8/6 |
L 8/7 |
blank |
- same for columns 5-8
5-8: VtpA primers
Thermal profile for qPCR:
- 7 minutes, 95C
- 10 seconds, 95C
- 30 seconds, 60C
- 30 seconds, 72C
- 1 minutes, 95C
- 30 seconds, 55C
- 30 seconds, 95C
- repeat for 40 cycles (from step 2)
Arbitrary expression = 10^(-(0.3012 * Ct) + 11.434)
qPCR issues and questions:
- Which samples are best to use? Vt water samples or Vt samples FROM oyster larvae
- downside to using oyster larvae samples for Vt is that RNA may be lacking
- Annealing temperature for 16S - normalizing? Emailed Elene, probably 50 - 55C??
- Which should I run next?
- Next to see if the 16S will work, for normalizing
- Tomorrow: run replicate samples again (24 total): plan to use different annealing temperature, replicates of Controls (no host present), 2 samples each of 380ppm of Vt with host present
- Hopefully I can get the normalizing gene down and then go from there
Next steps:
- Running Vt qPCR, with 16S and my 12 samples (in replicates) use 50C annealing temperature
- Need to set up new experiment, help Emma out
- Plan on answering one question at a time:
- focus on getting normalizing gene
- get some data focusing on one gene at a time with 4 hour and 24 hour post inoculation time points
- great to help out Emma for next experiment, could maybe process some samples after 24 hours, may also spill back into Seattle?!?
August 9, 2010
- Started working on proposal, scheduled in time with Emma to begin writing
- Extracted more RNA this weekend (in -80C)
- Emma and I created a schedule for the week
- Have a plan B if we have no amplification from today's qPCR!
Primers to use today:
Chitinase
VtpA: metalloprotease
VtpR: transcriptional regulator for metalloprotease and hemolysin secretion
qPCR results: (will download/export data soon)
- Amplification from all genes, no contamination
- next steps: focus on two genes tomorrow and answering one question
- Genes: 16S (normalizing) and VtpA (possible virulence factor)
- Question: Is there a difference in gene expression (metalloprotease) in Vt with and without a host present? Will be looking at samples from ambient CO2 from 4, 24, and 48 hours post-inoculation (and controls that correlate with treatment). Then will move on to another gene to examine (three total)
- Tomorrow: reverse transcribe the rest of the RNA samples for the next qPCR, if data is not sound, use Emma's RNA samples that include larvae. Possible that Vt virulence can be examined from the larvae RNA (got some amplification today from an oyster sample)
August 7, 2010
Took out 48 hour post inoculation sample from OA set-up
Spun down samples and took out last 1mL worth in microcentrifuge tubes and spun down more, most likely got a pellet today from Vt water
Also took 10mL of oyster larvae sample to look for Vt gene expression
- Finished RNA extractions (protocol below), will hopefully do qPCR tomorrow
- Did samples from 8/5 and 8/6 A, and one control at 380ppm and the control Vt culture, will run this with 3-4 primers
- should do calculations for this tomorrow
- qPCR sign up at 3pm tomorrow (Sunday)
- TO DO: work on proposal, paper, qPCR prep for tomorrow
RNA extraction protocol
- Spin off RNAlater
- Add Tris and mix/homogenize thoroughly, incubate 5 minutes at RT
- Add .2mL of chloroform and vortex a lot (pinkish), incubate
- Spin down and take off aqueous phase to new tube
- Add isopropanol (.5uL), mix and incubate
- Spin down 12X rpm for 8 minutes
- Wash pellet with 75% EtOH
- remove supernatant ("pellet" remains), let dry
- Need to remember where "pellet" would be
- add 20uL of DEPC H2O at the end, store in -80C freezer, continue reverse transcription tomorrow
REVERSE TRANSCRIPTION PROTOCOL
1. Mix your stock RNA sample by inverting tube several times.
2. Transfer 25ug of your RNA (.25ug of mRNA) to a fresh PCR tube. Bring the volume up to 5uL with PCR water. If necessary, spin tube briefly to pool liquid.
3. Incubate tube at 75C for 5mins in thermal cycler.
4. Transfer tube IMMEDIATELY to ice and incubate for at least 5mins.
5. Make Master Mix (MM)
PER RXN
4 ul 5x Buffer (AMV RT Buffer)
8 ul dNTPs (10 mM total)
1 ul AMV RTranscriptase
1 ul Oligo
dT
Primer
1 ul RNase free water
Total = 15 ul
- Add MM to tube with diluted RNA in it (total volume now 20 ul)
- Vortex
- Spot spin
- Incubate at RT for 10 min
- Incubate at 37C for 1 hr in thermocycler
- Heat inactivate @ 95C for 3 min
- Spot spin
- Leave cDNA on ice or store at –20C
qPCR Master Mix (uL): per reaction (for each MM today)
1. 2X Sybr MM 2.5 (25uL)
2. BSA 1.5 (15uL)
3. F primer 0.5 (5uL)
4. R primer 0.5 (5uL)
5. PCR H2O 8 (80 uL)
6. cDNA template 2
August 6, 2010
Took samples for Vt at 9:00AM
- pH and temperature were taken (update google doc)
- samples were not taken from G
- Need to make sure air is going to yellow line graduated cylinder
Last samples will be taken tomorrow morning (Saturday) 48 hours post-inoculation
Will take as much water (probably 3-4 50mL falcon tubes each) as possible in order to try to get a pellet after spinning down sample.
This weekend:
- Plan on starting RNA extractions (16) on Saturday
- Maybe finish up the other ones Sunday and Monday
- Try to start initial primer trials on Monday or Sunday; qPCR with primers and Vt with no host present
August 5, 2010
Protocol for Vt collection:
1. Supplies to OA lab: seriological pipette, 50mL falcon tubes (12? 24?), pen, parafilm
2. Start with uninoculated bottles and green tubes, wash out larvae, take larvae sample for counting and some for RNA later
3. No water samples collected from uninoculated bottles
4. collect water while inoculated bottles are being filtered, collect 1 or 2? 50mL falcon tubes from each
5. spin down tubes for 20 minutes, transfer to 1.5mL tubes and spin down again
5a. Streak on plate (quadrants) MA or TCBS, or take sample and dry/stain??
6. If two samples collected, put RNA later in one (50uL), or just put RNA later in the tube with pellet
7. Make sure waste goes into biohazard
8. bleach+waste solution+used tubes all go into bucket to sterilze
9. switch out bleach water for clean water and soak O/N
10. Make sure tubes are in -80C
11. Update lovely lab notebook and google doc spreadsheet
Issues
- Density change with controls... maybe don't add anymore water, just take?
- Yellow line controls (840ppm) cannot get air (ask Moose?) Temporarily have parafilm on top
- Significant differences between ambient and elevated CO2?
- make sure to swab and streak cleaned falcon tube for tomorrow.
- Inoculated with Vt today
- Took post-inoculation samples (duplicates, 1 is in RNA later)
- Post procedure, clean-up, needed supplies, where things are, problems...
- What's next?
- Share google doc with all data!
Two control graduated cylinders:
Planning on taking some water from sample and then replacing it (keep at approx 800mL)
Questions: is this the best way to sample?
Ways to wash falcon tubes?
Negative controls are correct?
supplies to take to Lab 6:
24 falcon tubes, sampling means: seriological pipette (50mL)
- Also need to follow up from presentation form last night!
August 4, 2010
RNA extraction protocol below
Helping Emma do about 24 samples a day of previously treated larvae.
Oyster Larvae, Vt and OA experiment
Set up:
- In Moose's lab with CO2 hook up
- Still static system, but maintained via CO2 water
- four larvae containers in approx 380ppm and four in approx 900ppm (readings will be taken)
- Two graduated cylinders are set up for Vibrio tubiashii controls
- Will plan on changing water, taking readings (CO2, salinity, pH)
Samples to take: (In addition to larvae samples for Emma)
Discarded water from larvae
Water samples from graduated cylinders
| CO2 |
Contents |
Type of sample |
Collect for: |
| 380ppm |
Vt |
control |
RNA, plate out |
| 900ppm |
Vt |
control |
RNA, plate out |
| 380ppm |
Vt+host |
treatment |
RNA, plate out |
| 900ppm |
Vt+host |
treatment |
RNA, plate out |
Collected for is subject to change as well!
*Duplicates will also be collected for each treatment/control (20 total)
Next step:
Growing up Vt tonight, will inoculate tomorrow, and do serial dilutions.
Vt from marine broth (probably one of diluted samples) will be taken for test run for RNA
Need to get all supplies and necessary things for RNA tonight
Primers:
chitinase, metalloprotease, 16S, VtpR - transcriptional regulator for metalloprotease + hemolysin secretion and swimming motility
August 3, 2010
Research question: What is the difference in genes are expressed by Vibrio tubiashii with a host present (oyster larvae) and host+Ocean acidification challenge? Will specifically be looking at virulence factors
- May be able to add a multiple species aspect...
Differential display
Differential Display Protocol
Made master mix for 7 reactions total, used for 5 reactions
3 cDNA samples and 2 blank water samples
Per reaction:
1uL cDNA
2uL 5mM arbitrary primer
1uL 10uM dtACP
6uL H2O
10uL See Amp2x Mix
19uL of reaction + 1 uL of cDNA = 20 uL total
We used primer #16
- Continue with differential display tomorrow
Armina status
Can take any Armina from Neuro lab. Will be mostly looking at bacterial ID of the lesions from the nudibranchs. Will be needing histology back first. This week, maybe look at publications on nudibranch histology to prep for examination of histo. Will get back maybe next Monday?
In lab:
Protocol:
Test for protease:
a. Centrifuge culture tubes at 26,00 rpm for 10 min at 4C
b. Remove 100 μl of supernatant and add to a microcentrifuge tube (discard the rest of the pellet and culture supernatant)
c. Add 400 μl of 1% azocasein
d. Incubate for 30 min at 37°C
e. Stop the reaction by adding 600 μl of 10% trichloric acetic acid (TCA)
f. Incubate on ice for 30 min
g. Centrifuged at 13,000 rpm for 5 min
h. Add 200 μl of 1.8 N NaOH to a new microcentrifuge tube
i. Add 800 μl of the reaction supernatant to the tube with the NaOH
The supernatant will turn an extremely bright and obvious orange upon a positive result for the protease, it will remain whatever color the supernatant was (not always clear, might have slight yellowish tint) if it is negative for the protease
Azocasein Protease Assay use in detecting pathogenic Vibrio spp. Reference:
Hasegawa, H, Lind, E.J., Boin, M.A., Hase, C.C.
The Extracellular Metalloprotease of Vibrio tubiashii Is a Major Virulence Factor for Pacific Oyster (Crassostrea gigas) Larvae. Applied and Environmental Microbiology, July 2008, p. 4101-4110, Vol. 74, No. 13.
Future project:
Objectives:
- Bacterial ID of lesions on nudibranch
- look at their histology and other tests
- Look at Vibrio challenge from oyster larvae and possibly anemone and nudibranch
- Taking sample Vibrio RNA before and after infection
- Talk to Elene today about how to extract Vibrio samples from challenged organisms
- most likely will be from surrounding water, and that will be pathogen with host presence
August 2, 2010
Oyster Larvae:
Have been helping Emma out with oyster larvae project. This past weekend was changing out oyster larvae water and feeding them algae. Larvae came from Taylor Shellfish hatchery. They were challenged yesterday (August 1st) with Vibrio tubiashii. Game plan
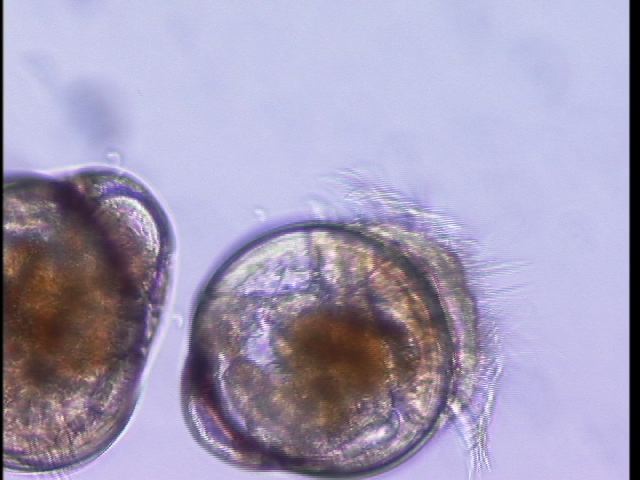
Oyster serial dilutions were performed:
plate counts of 167 and 144 at 10E-6 dilution
Therefore our oyster larvae
Armina
- Two types of lesions now
- scrape and gram stain both (need to find sterile means)
- gross examination of both types of lesions
- hopefully histology for both (need to send out tomorrow)
1. New lesion does not look like older one. It is "bumpy" and does not look as necrotic as the previously found one. Among the bumpy texture of the lesion there are small black bits?? sort of long? Will need to get a better picture/closer look to describe further
2. Older lesion has flatter texture
This week:
- Plan on obtaining more samples from Neuro class
- Sending out New group lesion and old group lesion for histology
- samples from histo will also be kept in molecular EtOH for further analysis
- Differential gene expression between infected and healthy individuals (will get healthy ones from Neuro class)
Serum Agglutination Test
a. Add 50 uL of sterile seawater and 25 uL of your culture to one slide (control). What would be a good additional control here?
b. To one slide (your test slide) add 25 uL of sterile seawater and 25 uL of your culture and 25uL of the thawed polyclonal
antibody.
c. Gently mix solutions on both slides with individual pipette tipes (gently suck up and down)
d. Incubate at room temperature for 5 mins
e. View at 10x or 4x on the scope to compare degree of agglutination of the cells.
July 31, 2010
Oyster Larvae
Counting larvae, feeding, and changing their water everyday.
Making marine broth today to grow up Vt so challenges can start on Monday
Projects
- Will be finalizing plans and figuring out group logistics
- Need to order things/bring from SAFS by Monday afternoon (will be brought up Tuesday)
- Emailed Elene about bringing up all available V. tubiashii primers
- Still looking for appropriate primers and approach for rickettsia in Armina californicus
Next up:
Start working on Quiz 2 this weekend: ISH troubleshooting
- can refer to below notes from July 28th entry
July 30, 2010
Brainstorming all day
Two project ideas:
1) Look at Armina califoricus stress response to rickettsia
2) Many animals in the lab will be challenged with Vibrio tubiashii, I would like to compare/contrast V. tubiashii expression and products via qPCR in different species. Not sure if I will find anything
Challenges:
- Figuring out samples
- finding primers! (literature or online)
- timing/logistics
- qPCR (maybe try to run a trial qPCR in the next few days with some kind of positive and negative control)
Primer tutorial
Initial tips/ideas:
- Consensus sequence: bias come up when aligning. Program will call it at certain percentages
- Finding conserved genes
- Need to figure out alignment programs in order to make more general sequences that will amplify with any trematode species
- Along the same lines, maybe still use my approach (shu251) but do more research and observe more species in the area to determine more specific species and go from there
- Look at EST for wanted sequence, BLAST it and see what comes up
- BLASTX (nucleotide and then translate to all possible) - consider that proteins are more conserved overall
- tBLASTX = nucleotide translating in all six reading frames and doing it again and then comparing all options
- Need to select (non-human) and EST
- Next steps: Next time design 3-5 primer sets. I would prefer to use both literature and something like BLAST or Primer3
- Do research for specific specific, universal primers
- Possibly examine genes that may be up regulated during stress
How do I use Primer 3?????
- Use GenBank to find sequence, chose as many as possible
- Save the sequences in FASTA format on computer
- Use ClustalW2 to align and find consensus sequence (resource)
- Input FASTA format to Primer3 or BLAST program
- Find primers and try out in Amplify3
Amplify 3 (virtual PCR, test primers)
- Download
- Import sequence from GenBank
- Copy sequence and go to file--> open target file --> paste in sequence
- Format sequence
- Put in primers (other window) --> check boxes of wanted primers
- Go to PCR, amplify
- Show....
- Will show potential PCR product
Re-do Western Blot
- Steven re-did Western blot last night (thanks!)
- Stained it this morning, will see results (hopefully) in approx. 1 hour
- Results: did not work, due to antibody not binding with anemone
July 29, 2010
Dissection of Ostrea edulis
- Insert knife into hinge and cut adductor muscle
- Open up oyster
- Able to extract haemolymph for slide and imprint heart onto another slide
- Careful examination of entire anatomy of oyster (images)
- Looking for Bonamia in haemolymph
- Example: Arrows point to Bonamia in haemolymph
- No Bonamia detected in my sample, but did see lots of haemolymph
Follow up of protein extraction and gel
- Our sample was much more concentrated than other samples (2nd from right well- pictured below)
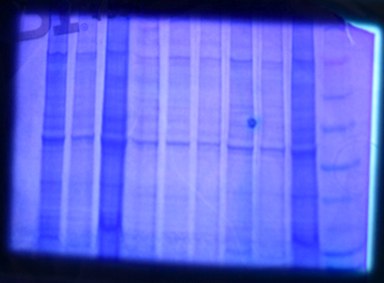

- Diluted 25uL of protein extract with 75uL of CellLytic buffer; this way we can still add 30uL on new gel
- Similar concentrations of sample are needed for the Western Blot procedure
New gel: loading 30uL on one half and 20uL on other half. Will cut gel in half, one side for visualization and one side for transfer.
Western Blot background
Gel run with SDS, which is negatively charged. A charge will be applied to the gel so the proteins will migrate out of the gel onto a positively charged membrane, nitrocellulose membrane.
General Western Blot procedure:
- Block membrane to reduce unwanted proteins
- Add primary antibody with membrane. Primary antibody is chosen to be specific for the protein that we are trying to isolate
- Rinse membrane so that unwanted primary antibody is removed. The Secondary antibody is then added to the membrane. This is usually species-specific.
- Rinse membrane again to remove unnecessary secondary antibody. An enzyme is added for detection; usually there will be a visual emission of light or change of intensity of the band.
- Transfer proteins to the membrane!
Western Blot protocol day 1:
1. Soaking nitrocellulose membrane in blocking solution on shaker for approx 15 minutes, decant
2. Prep ponceau dye to visualize the membrane
3. Rinse membrane with water and then decant, repeat
4. Make primary antibody solution
5. Incubate membrane with the antibody solution, recommended overnight, but we did not do this
6. Wash membrane with antibody wash, decant, repeat x3
7. Incubate membrane in secondary antibody solution, decant
8. Rinse membrane with water, decant, repeat x2
9. Apply poceau dye.. magic!
Result: No magic yet, should see bands appear within 1 hour.
Looking forward to projects, considering collaboration with larval course for samples. Maybe challenge samples with pathogen and CO2. Another consideration is the nudibranch armina rickettsia
The previous thought of using Pisaster ochraceus and wasting disease will not work due to lack of samples, but will try to pursue finding infected sea stars on the island (next low tide: August 5th)
July 28, 2010
Day 3: ISH
Last few steps of ISH protocol
Probe did not seem to work. We should have seen general tissue to be stained a light brown, pale color and rickettsia stained a dark purple/almost black. Probe stained something a dark color, but it was in incorrect location and shape for the rickettsia-like pathogen region.
- First we examined the H&E slides of our same samples (with what kinds of rickettsia should be present):
- 08: 30-3; classic, new, and stippled
- 08:1-7; new
- Classic: homogenous pattern "egg shape", stained darker
- New: "Cracked" egg shape, still a dark stain
- Stipple: Other? Lighter stain and less defined shape?
- Our ISH stained slides did not work in the end (probe did not work)
- How to troubleshoot probe for in situ hybridization?
- Probe may not have worked for various reasons:
- Denaturing or annealing temperatures were not optimal
- Storing of probe or possibly incubation periods with probes did not work
- Temperature and length of washes and incubation may have needed to be optimized
Protein extraction
Background: Anemone heat stress (29C for approx: 2 hours). Control are in the sea water tanks in lab.
Three anemones for each treatment.
Species: Anthopleura elegantissima
Protein extraction protocol
1. Cut anemone and get approx 25mg, and use razor blade and pestel to homogenize appropriately
- Matthew and I have a heat shocked anemone
- weight for extraction was 0.06g
- added a total of 800uL of CellLytic solution
- Label: HS_M&S_protein
- stored in fridge during lunch...
4. put in water bath for 5-10 minutes, spin down
5. load gel (buffer for gel is 1X SDS buffer)
6. Gel run for 45 minutes at 115V
- final gel image will give us an idea of the quality of protein we have, the amount and possibly some of the different times (size depending)
July 27, 2010
Day 2: ISH
Still following protocol.
- Slides with coverslips have been sitting in 53C humid chamber overnight
- Will perform a series of stringency washes (Protocol)
- Last step is to block the tissues with "Blocking Buffer" at Room temperature for an hour (go to lunch)
- Detection! (tomorrow)
Next steps:
Saw no development after 15 minutes so we are incubating overnight at room temperature.
Tomorrow we will be rinsing with dH2O, counterstaining with 0.05% Bismark Brown Y and rinsing again with dH2O. Then a series of EtOH rinses and then after air drying we will permount the slides and view!
Group project brainstorming:
Initial ideas:
- Pisaster ochraceus wasting disease (histo and maybe some microbial community analysis?)
- Antimicrobial peptides and innate immunity responses (possibly try to collaborate with larval class)
- Still figuring out logistics of sampling and techniques for evaluating Pisaster ochraceus.
- possibly able to go sampling tomorrow during lunch hour? Will also be considering all options
July 26, 2010
Withering Syndrome ISH
Day 1:
Protocol
General:
1. Tissue deparaffinization; xylene and ethanol (be careful with xylene!)
2. Permeabilization of tissues
3. Prehybridization
4. Incubate in prehybridization buffer for 1-1.5 hr at 37C
5. rinse in 2xSSC and briefy dry
6. add probe to prehybridization buffer (1:373 concentration)
7. add this (300 uL) to tissue sections (300uL each)hybridization: 30 minutes
8. cover with DNAse-free coverslips
9. Incubate overnight
*follow above link for protocol, be aware of time management
PCR incorporates the sequence into probe (labeled dye).
If similar sequence on slide, denaturation and annealing will cause the dye to stick to that tissue.
- Withering syndrome affects black and red abalone. It is most prevalent off the coast of California.
- The bacterium is abalone rickettsia, Candidatus Xenhaliotis californiensis
- Pathogen is found in digestive tract of abalone.

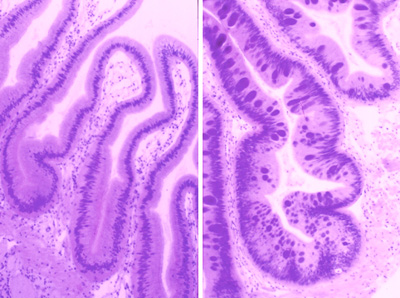
ABOVE: Left is normal digestive tract and the right image is tissue from withered red abalone.
Designing primers over weekend:
Two effective methods:
1. Use published work to find previously used primers
2. Use NCBI and BLAST to find sequences and plug into alignment software, such as Geneious
To design primers I first scanned NCBI starting with trematodes. Then I began looking a the more specific lineage: Cercaria.
With all Cercaria species in mind, I chose the cytochrome oxidase subunit 1 gene to look for primers. I also scanned the other species of Cercaria to see if a cytochrome oxidase subunit 1 specific primer would possibly work with other sequences available on NCBI. I began with one species Cercaria batillaria and ran the accession ID in the PrimerBLAST program on NCBI.
This program allows a check for specificit, where I put in Cercaria, this way the primers may be designed to detect the cytochrome oxidase subunit 1 in all Cercaria.
My results came up with 6 primer pairs:
Option 1:
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
TGGCGCATTGAGGGGCTTCG |
Plus |
20 |
473 |
492 |
60.04 |
65.00% |
| Reverse primer |
TGGGGTACAGGCAAGGTAGCCA |
Minus |
22 |
746 |
725 |
59.25 |
59.09% |
| Internal oligo |
Plus |
||||||
| Product length |
274 |
||||||
Option 2:
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
ATCGGGGTTCCGACGGGCAT |
Plus |
20 |
172 |
191 |
60.32 |
65.00% |
| Reverse primer |
GGGGTACAGGCAAGGTAGCCA |
Minus |
21 |
745 |
725 |
57.86 |
61.90% |
| Internal oligo |
Plus |
||||||
| Product length |
574 |
||||||
I chose these two primer pairs (of six) because the product lengths differ enough to possibly examine both on the same gel and differentiate. However, the specificity and effectiveness may be different between the two primer options.
Resources
Next steps:
Would like to examine more methods for finding primers. Will be looking into ClustarX online for alignment of sequences and looking at Geneious as another option.
July 24, 2010
Analyzed qPCR results today.
Protocol is to find the ct (cycle threshold) values and use arbitrary expression formula to get the expressed value. With our two primers, Actina and Cjunk, we will be dividing Cjunk by actin expression values to get a normalized number. This division can be done because, theoretically, Cjunk is a housekeeping gene and actin may have elevated values with the presence of the trematodes. The arbitrary expression values can be used for relative comparisons.
Arbitrary expression = 10^(-(0.3012 * Ct) + 11.434)
My results came up all negative; no expression, too low to detect, or something wrong with my reactions. In contrast to the rest of the class, I used solely the PCR water, whereas they used the nanopure water. Thus there is a possible source of contamination. However, with no expression in any of my reactions, there is most likely something wrong with my master mix or there is some sort of inhibition.
Future qPCR:
- try to use Lab 10!
- use own Master Mix reagents and do it all at one time (difficult to share, there were a few times I had to use multiple aliquots because I needed to find more in the lab or the correct pipettes were not available).
- possibly try two types of master mix to make sure one is not contaminated
- prepare own primer dilutions
- future controls: negative, positive, RNA (for gen. DNA), if available- two MM.
Next steps:
- try to DNAse
- possible new extraction?
- run RNA with qPCR to see if genomic DNA is present
July 23, 2010
Following up with RNA extraction
Reverse transcription and set-up for qPCR:
1. Thaw on ice and mix RNA stock (keep on ice!), set up heat blocks for 75C, 42C, 95C
2. heat block for 5 min. (70C)
3. Transfer to ice for approx. 10 minutes --> cDNA
4. Prep master mix (total for each rxn will be 7.25 uL of MM and .75 uL of cDNA)
5. run rxn for 1 hour at 42C and 3 min. at 95C (inactivation)
6. amplified cDNA!
Follow-up:
Re-streaking nudibranch NL site, on both marine agar and TCBS plates: suspected that bleaching and rising method for streak plates was NOT effective. Today loops and flame sterilization is available.
Along with re-streaking NL sites for isolation, will also streak eel grass cultures to see what bacteria we can see from that.
Gram staining: All gram negative rods
qPCR for cDNA (from RNA reverse transcription)
Two samples of cDNA (stored at -4C)
- SH2
- AMM1
Need duplicates of qPCR reactions and negative controls, two separate master mixes because we are using two primers.
Primers:
1. "Actin": Ll_actin_592-611_F
Ll_actin_761-742_R
2. "c-junk": Ll_cjunk_203-184_R
Ll_cjunk_871-106_F
Need to multiply MM components by 10 because each sample needs enough for 5 reactions.
MM1: actin and MM2: c-junk
qPCR Master Mix (uL): per reaction (for each MM today)
1. 2X Sybr MM 2.5 (25uL)
2. BSA 1.5 (15uL)
3. F primer 0.5 (5uL)
4. R primer 0.5 (5uL)
5. PCR H2O 8 (80 uL)
6. cDNA template 2
Template on well plate
| MM1 |
MM1 |
MM1 |
MM2 |
MM2 |
MM2 |
MM1 |
MM1 |
MM1 |
MM2 |
MM2 |
MM2 |
| SH2 |
SH2 |
neg |
SH2 |
SH2 |
neg |
AMM1 |
AMM1 |
neg |
AMM1 |
AMM1 |
neg |
| actin |
actin |
actin |
cjunk |
cjunk |
cjunk |
actin |
actin |
actin |
cjunk |
cjunk |
cjunk |
July 22, 2010
Follow-up:
Plates from NL and L sites
NL plates did not show very many colonies, some small white colonies on Marine agar. Matthew's plate contained both green and yellow colonies on the TCBS plates
Lesion sites had yellow colonies on TCBS plates and small white colonies on marine agar
Hopefully gram stain colonies tomorrow (7/23)
RNA Extraction
(protocol from lab handout)
Isolation:
1. Turn on heating block to 55C. Also turn on spectrophotometer.
2. Add 500uL of TriReagent to a 1.5mL snap cap tube. Store on ice.
3. Cut a piece of frozen tissue weighing between 50-100mg and add to tube containing TriReagent.
4. Carefully homogenize the tissue using a disposable pestle.
5. Add an additional 500uL of TriReagent to the tube and close the tube.
6. Vortex vigorously for 15s.
7. Incubate tube at room temperature (RT) for 5 mins.
8. In the fume hood, add 200uL of chloroform to your sample and close the tube.
NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
9. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
10. Incubate tube at RT for 5 mins.
11. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
12. Gently remove tube from microfuge. Be sure not to disturb the tube.
13. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
14. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
15. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
16. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
17. Incubate at RT for 10 mins.
18. Spin in refrigerated microfuge at max speed for 8 mins.
19. A small, white pellet (RNA and salts) should be present. If not, do not fret. Continue with procedure.
20. Remove supernatant.
21. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
22. Spin in refrigerated microfuge at 7500g for 5mins.
23. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
24. Briefly spin tube (~15s) to pool residual EtOH.
25. Using a small bore pipette tip (P20 or P200 tips), remove remaining EtOH.
26. Leave tube open and allow pellet to dry at RT for no more than 5mins.
27. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
28. Incubated tube at 55C for 5mins. to help solubilize RNA.
29. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
30. Quantitate RNA yield using spectrophotometer (we will be using the Nanodrop).
RNA extraction notes:
Had two tubes:
yellow: AMM1 and blue: SH2
- hopefully both have RNA!
Notes that AMM1 pellet took longer to resuspend after adding the DEPC H2O, had to heat for 5-10 minutes and vortex frequently.
Tomorrow, we will quantify RNA
Eel grass collected this morning
Two bags of eel grass, one is healthy, one appears black/white/brown is places.
Procedure for growing up cultures:
1. Wash eel grass in beta dye
2. wash in sterile sea water
3. cut into many pieces to increase surface area and then place in QPX media
- 6 different eel grass blades in 6 well plates
- to compare, eel grass blades were also cut and placed on marine agar and TCBS agar
July 21, 2010
Examination of arminia semperi (maybe semperi?)
Obtained sampled from neuro class at FHL. Nudibranch have lesions on epidermis. Small lesions that break up their white to orange/brown colored stripes. Non-healthy arminia have behavior differences from healthy ones. Neuro class has separated healthy from non-healthy, but would still like to observe the arminia from the lab set up.
Background: Arminia were collected near Tacoma, Washington and there is obvious behavior and coloration changes in the unhealthy animals.
Procedure:
We streaked out both lesion sites and non-lesion sites. Streaked on TCBS and marine agar plates.
Labels: L1, L2, and L3 are Lesion samples and NL1, NL2, NL3 are non-lesion sites.
For further analysis we scraped samples from lesion and non-lesion sites and gram stained them. Further analysis is needed. Hopefully tomorrow we can obtain more samples and relax the nudibranch. Lesion and sites were difficult to extract tissue from because the animal kept contracting.
Observations:
Gram negative rods were seen in lesions sites and "amoebe-like" organisms were seen on the lesion site slides as well. They were cilliated and move very quickly (stained purple)
When scraping the nudibranch, the lesion sites were easier to get tissue from, particles were easily taken off, whereas the non-lesion sites were more difficult to get scraped samples from.
Dissected infected and uninfected nudibranchs; did not see any apparent differences of internal organs.
PART II:
DNA extraction:
Used QIAGEN stool kit (link)
Sample ID: SH2
SH2 sample was L. scutulata and infected
Littorina qPCR method/master mix:
(in uL)
Promega GoTaq 12.5
BSA 1.5
F primer 1
R primer 1
dH2O 7
DNA template 2
- used buffer??
July 20, 2010
Histology
Examined multiple histology slides from various species (abalone, mussel, etc.)
Observations:
- based on red abalone
Muscle: fibrous appearance, uniform in color, larger masses
Digestive gland: (image in notebook) darker color
Intestine: associated with blue-ish coloration near-by
Rectum: similar to intestine shape, but wider membrane, and frilly appearance, structure is also tighter
Nerves: uniform in color, 'matte' appearance
Gills: looked like gill filaments!
Notes:
Some histological slides are difficult to make inferences from due to histology preparation
Will be going into lab to examine more histological stains and begin comparing them to diseased tissue
Littorina Dissection
Procedure:
1. crack open shell in order to remove shell
2. remove all shell pieces, "massage" animal out of shell or make certain to crack the top of shell where columellar muscle is
3. once all shell is removed examine digestive gland for infection and cut off
4. remove operculum
5. slide up hood from head and remove head
6. save some foot tissure for DNA extraction
7. put rest of specimen in tube for RNA
8. freeze at -20 C
Notes:
Dissection of L. sitkana and L. scutulata
Dissected 3 L. sitkana and 2 L. scutulata
Both scutulata were infected and images of trematodes can be found here
Observed bright blue pigment behind head of L. scutulata that was very sick
*note to self electronic lab notebook will be deleted when using 'fn' "home" buttons on Mac.
July 19, 2010
Dissections
Cockle: used other bivalve anatomy diagrams as reference
Gastropod: Difficult to crack open shell to remove columellar muscle, found and observed eggs from gastropod (post image later)
On top of recalling my FISH 310 notes, use of some great online references for invertebrate anatomy will be useful:
http://webs.lander.edu/rsfox/invertebrates/
https://catalysttools.washington.edu/workspace/sr320/4/60314